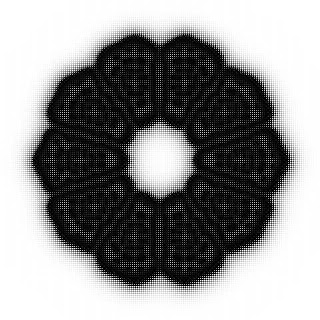
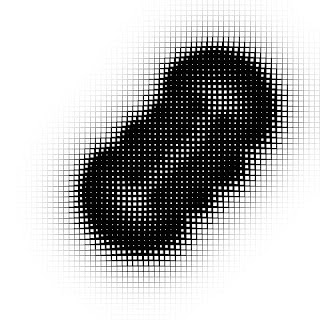
Mastering Turing Patterns: A Guide to Reaction-Diffusion in p5.js and Processing
This article explores creative coding with p5.js and Processing to generate organic-looking, intriguing Turing patterns.
This article covers the essential concepts for creating captivating visuals, provide simple example code, and share an advanced application. With these tools, you'll be able to create your own original work.
Turing Patterns: An Overview
Simply put, Turing patterns resemble the intricate patterns on tropical fish or leopards fur. These patterns emerge from a mathematical process known as a reaction-diffusion system.
English mathematician Alan Turing originally developed the theory behind these systems to explain how patterns in nature form.
Wikipedia : Turing pattern
Generating Turing patterns requires intensive computation—exactly where computers excel.
Basic p5.js Implementation
The following p5.js code demonstrates the fundamentals of drawing a Turing pattern using the Gray-Scott model.
/*
* Reaction-Diffusion system by the Gray-Scott Model.
* basic example.
*
* @author @deconbatch
* @version 0.1
* p5.js 1.1.3
* license CC0
* created 2022.03.26
*/
const w = 480;
const h = w;
const cSiz = 3; // cell size
const pCnt = 500; // calculation count
function setup() {
createCanvas(w, h);
noLoop();
const lab = new Labo(cSiz);
lab.init();
for (let i = 0; i < pCnt; i++) {
lab.proceed();
}
lab.observe();
}
/*
* Labo : reaction-diffusion system.
*/
class Labo {
cellSize;
matrixW;
matrixH;
diffU;
diffV;
cells;
constructor(_cSiz) {
this.cellSize = _cSiz;
this.matrixW = floor(width / this.cellSize);
this.matrixH = floor(height / this.cellSize);
this.diffU = 0.9;
this.diffV = 0.1;
this.cells = new Array();
}
/*
* init : initialize reaction-diffusion system.
*/
init() {
for (let x = 0; x < this.matrixW; x++) {
this.cells[x] = [];
for (let y = 0; y < this.matrixH; y++) {
this.cells[x][y] = new Cell(
map(x, 0.0, this.matrixW, 0.03, 0.12), // feed
map(y, 0.0, this.matrixH, 0.045, 0.055), // kill
1, // u
(random(1) < 0.1) ? 1 : 0 // v
);
}
}
}
/*
* proceed : proceed reaction-diffusion calculation.
*/
proceed() {
// calculate Laplacian
const nD = Array(); // neighbors on diagonal
const nH = Array(); // neighbors on vertical and horizontal
for (let x = 0; x < this.matrixW; x++) {
for (let y = 0; y < this.matrixH; y++) {
// set neighbors
nD[0] = this.cells[max(x-1,0)][max(y-1,0)];
nD[1] = this.cells[max(x-1,0)][min(y+1,this.matrixH-1)];
nD[2] = this.cells[min(x+1,this.matrixW-1)][max(y-1,0)];
nD[3] = this.cells[min(x+1,this.matrixW-1)][min(y+1,this.matrixH-1)];
nH[0] = this.cells[max(x-1,0)][y];
nH[1] = this.cells[x][max(y-1,0)];
nH[2] = this.cells[x][min(y+1,this.matrixH-1)];
nH[3] = this.cells[min(x+1,this.matrixW-1)][y];
// Laplacian
let c = this.cells[x][y];
let sum = 0.0;
for (let i = 0; i < 4; i++) {
sum += nD[i].valU * 0.05 + nH[i].valU * 0.2;
}
sum -= c.valU;
c.lapU = sum;
sum = 0.0;
for (let i = 0; i < 4; i++) {
sum += nD[i].valV * 0.05 + nH[i].valV * 0.2;
}
sum -= c.valV;
c.lapV = sum;
}
}
// reaction-diffusion
for (let x = 0; x < this.matrixW; x++) {
for (let y = 0; y < this.matrixH; y++) {
let c = this.cells[x][y];
let reaction = c.valU * c.valV * c.valV;
let inflow = c.feed * (1.0 - c.valU);
let outflow = (c.feed + c.kill) * c.valV;
c.valU = c.valU + this.diffU * c.lapU - reaction + inflow;
c.valV = c.valV + this.diffV * c.lapV + reaction - outflow;
c.standardization();
}
}
}
/*
* observe : display the result.
*/
observe() {
background(0);
fill(255);
noStroke();
for (let x = 0; x < this.matrixW; x++) {
for (let y = 0; y < this.matrixH; y++) {
let cx = x * this.cellSize;
let cy = y * this.cellSize;
let cs = this.cells[x][y].valU * this.cellSize;
rect(cx, cy, cs, cs);
}
}
}
}
/*
* Cell : holds cell informations.
*/
class Cell {
feed;
kill;
valU;
valV;
lapU;
lapV;
constructor(_f, _k, _u, _v) {
this.feed = _f;
this.kill = _k;
this.valU = _u;
this.valV = _v;
this.lapU = 0;
this.lapV = 0;
}
standardization() {
this.valU = constrain(this.valU, 0, 1);
this.valV = constrain(this.valV, 0, 1);
}
}
Note: If you're running this on OpenProcessing, be mindful of "loop protection" settings due to the high computation time.
This implementation uses the Gray-Scott model. The resulting pattern evolves based on the reaction-diffusion parameters mapped across the XY axes.
Code Overview
The Labo class handles reaction-diffusion calculations. The calculation scope is 'matrixW x matrixH' area. The Cell class represents a grid square in the matrix. The cell[matrixW][matrixH] array represents all cells in the computational grid.
'class Labo init()' method initializes all cells in the computational grid. 'class Labo proceed()' calculates the reaction-diffusion equations once. The number of iterations is configurable.
This process is computationally expensive and can take time to render. The calculation time is proportional to pCnt and increases fourfold when cSize is halved.
With these parameters, execution took about one minute on my machine.
const cSiz = 3; // cell size
const pCnt = 500; // calculation count
The two-dimensional array this.cells[x][y] holds the calculation results.
While this example generates monochrome images, you can customize the output as desired.
Tips for Creating Captivating Patterns
1.Tweak the Parameter Values
The visual characteristics depend entirely on the parameters within the reaction-diffusion equations.
The example code uses four parameters. It sets diffU and diffV to constant values while varying feed and kill based on XY coordinates.
// diffU, diffV
this.diffU = 0.9;
this.diffV = 0.1;
// feed, kill
this.cells[x][y] = new Cell(
map(x, 0.0, this.matrixW, 0.03, 0.12), // feed
map(y, 0.0, this.matrixH, 0.045, 0.055), // kill
For example, setting feed and kill to values A and B produces these characteristic patterns:
The example code will create the patterns when the values of 'feed' and 'kill' values fall within these ranges:
feed : 0.03 - 0.12
kill : 0.045 - 0.055
You can change the values of 'diffU' and 'diffV'. However, finding interesting patterns with these parameters can be challenging.
Seeding the Pattern
The initial state of the "V" values acts as the seed for your pattern. While the basic example code randomly sets some V values to 1, you can achieve specific structures by manually setting seeds in Labo.init().
this.cells[x][y] = new Cell(
(random(1) < 0.1) ? 1 : 0 // v
You can alter the resulting pattern by changing how V values are initialized.
Try setting a single point in the center, or arranging seeds in a circular formation to see how the pattern radiates outwards.
// Example to set circular.
for (let t = 0; t < TWO_PI; t += PI * 0.2) {
let x = floor(this.matrixW * (0.5 + 0.25 * cos(t)));
let y = floor(this.matrixW * (0.5 + 0.25 * sin(t)));
this.cells[x][y].valV = 1;
}
Resolution vs. Iteration
The relationship between the cell size (cSiz) and the iteration count (pCnt) drastically affects pattern appearance. Smaller cells provide higher resolution but require many more iterations to "grow" the pattern.
const cSiz = 3; // cell size
const pCnt = 1000; // calculation count
Creative Coding: Advanced Applications
Below is an advanced application using the concepts discussed. These examples are shared under the CC0 license.
The 'p5.js' application example code.
Change 'class Labo init()' below.
/*
* init : initialize reaction-diffusion system.
*/
init() {
const hW = floor(this.matrixW * 0.5);
const hH = floor(this.matrixH * 0.5);
for (let x = 0; x < this.matrixW; x++) {
this.cells[x] = [];
for (let y = 0; y < this.matrixH; y++) {
let d = dist(x, y, hW, hH);
let f = map(sin(TWO_PI * d * 3 / hW), -1, 1, 0.12, 0.03);
this.cells[x][y] = new Cell(
f, // feed
0.045, // kill
1, // u
0 // v
);
}
}
for (let t = 0; t < TWO_PI; t += PI * 0.2) {
for (let r = 0.1; r < 0.4; r += 0.1) {
let x = floor(this.matrixW * (0.5 + r * cos(t)));
let y = floor(this.matrixW * (0.5 + r * sin(t)));
this.cells[x][y].valV = 1;
}
}
}
The 'Processing' application example code.
It's the same application example written in 'Processing'.
/**
* Reaction-Diffusion system by the Gray-Scott Model.
* application example.
*
* @author @deconbatch
* @version 0.1
* Processing 3.5.3
* license CC0
* created 2022.03.26
*/
void setup() {
size(480, 480);
noLoop();
int cSiz = 2; // cell size
int pCnt = 1000; // calculation count
Labo lab = new Labo(cSiz);
lab.init();
for (int i = 0; i < pCnt; i++) {
lab.proceed();
}
lab.observe();
}
/*
* Labo : reaction-diffusion system.
*/
public class Labo {
int cellSize;
int matrixW;
int matrixH;
float diffU;
float diffV;
Cell[][] cells;
Labo(int _cSiz) {
cellSize = _cSiz;
matrixW = floor(width / cellSize);
matrixH = floor(height / cellSize);
diffU = 0.9;
diffV = 0.1;
cells = new Cell[matrixW][matrixH];
}
/*
* init : initialize reaction-diffusion system.
*/
void init() {
float hW = matrixW * 0.5;
float hH = matrixH * 0.5;
for (int x = 0; x < matrixW; x++) {
for (int y = 0; y < matrixH; y++) {
float d = dist(x, y, hW, hH);
float f = map(sin(TWO_PI * d * 3.0 / hW), -1.0, 1.0, 0.03, 0.12);
cells[x][y] = new Cell(
f, // feed
0.045, // kill
1.0, // u
0.0 // v
);
}
}
for (float t = 0.0; t < TWO_PI; t += PI * 0.2) {
for (float r = 0.1; r < 0.4; r += 0.1) {
int x = floor(matrixW * (0.5 + r * cos(t)));
int y = floor(matrixW * (0.5 + r * sin(t)));
cells[x][y].setV(1.0);
}
}
}
/*
* proceed : proceed reaction-diffusion calculation.
*/
void proceed() {
for (int x = 0; x < matrixW; x++) {
for (int y = 0; y < matrixH; y++) {
// neighbors on diagonal
Cell[] nD = new Cell[4];
nD[0] = cells[max(x-1,0)][max(y-1,0)];
nD[1] = cells[max(x-1,0)][min(y+1,matrixH-1)];
nD[2] = cells[min(x+1,matrixW-1)][max(y-1,0)];
nD[3] = cells[min(x+1,matrixW-1)][min(y+1,matrixH-1)];
// neighbors on vertical and horizontal
Cell[] nH = new Cell[4];
nH[0] = cells[max(x-1,0)][y];
nH[1] = cells[x][max(y-1,0)];
nH[2] = cells[x][min(y+1,matrixH-1)];
nH[3] = cells[min(x+1,matrixW-1)][y];
// lapU
Cell c = cells[x][y];
float sum = 0.0;
for (int i = 0; i < 4; i++) {
sum += nD[i].getU() * 0.05 + nH[i].getU() * 0.2;
}
sum -= c.getU();
c.setLapU(sum);
// lapV
sum = 0.0;
for (int i = 0; i < 4; i++) {
sum += nD[i].getV() * 0.05 + nH[i].getV() * 0.2;;
}
sum -= c.getV();
c.setLapV(sum);
}
}
// reaction-diffusion
for (int x = 0; x < matrixW; x++) {
for (int y = 0; y < matrixH; y++) {
Cell c = cells[x][y];
float reaction = c.getU() * c.getV() * c.getV();
float inflow = c.getFeed() * (1.0 - c.getU());
float outflow = (c.getFeed() + c.getKill()) * c.getV();
c.setU(c.getU() + diffU * c.getLapU() - reaction + inflow);
c.setV(c.getV() + diffV * c.getLapV() + reaction - outflow);
c.standardization();
}
}
}
/*
* observe : display the result.
*/
void observe() {
background(0);
fill(255);
noStroke();
for (int x = 0; x < matrixW; x++) {
for (int y = 0; y < matrixH; y++) {
int cx = x * cellSize;
int cy = y * cellSize;
float cs = cells[x][y].getU() * cellSize;
rect(cx, cy, cs, cs);
}
}
}
}
/**
* Cell : hold the information about each cell.
*/
public class Cell {
private float feed;
private float kill;
private float valU;
private float valV;
private float lapU;
private float lapV;
Cell(float _f, float _k, float _u, float _v) {
feed = _f;
kill = _k;
valU = _u;
valV = _v;
lapU = 0.0;
lapV = 0.0;
}
public void setLapU(float _l) {
lapU = _l;
}
public void setLapV(float _l) {
lapV = _l;
}
public void setU(float _u) {
valU = _u;
}
public void setV(float _v) {
valV = _v;
}
public float getFeed() {
return feed;
}
public float getKill() {
return kill;
}
public float getU() {
return valU;
}
public float getV() {
return valV;
}
public float getLapU() {
return lapU;
}
public float getLapV() {
return lapV;
}
public void standardization() {
valU = constrain(valU, 0.0, 1.0);
valV = constrain(valV, 0.0, 1.0);
}
}